I had several conversations at the INSNA conference that made me realize it might helpful to blog some really short examples of lesser-known features and hard-to-remember tricks for the R packages network, networkDynamic, statnet, etc.
This example was prompted by a question on the statnet help list:
Multiplex nets
Even people who use statnet a lot often don’t know that Carter Butts designed the network data structure to support “multiplex” ties. Also known as “parallel edges” or “different tie types in the same network”. Multiplexity means that that there can be more than one (directed) edge between a pair of vertices. At the moment, multiplexity is only supported in that the package doesn’t prevent you from accidentally adding an extra edge between vertices, and the network object has flag that can be set to tell other functions “watch out, this network can have multiple ties”.
> library(network)
# create a network, setting the multiplex flag
> multi< -network.initialize(5,multiple=TRUE)
> is.multiplex(multi)
[1] TRUE
# add parallel edges between v1 and v2
# also add an attribute named 'type' to each
> multi< -add.edges(multi,tail=c(1,1,1,1),head=c(2,2,3,4),
names.eval='type',vals.eval=c("A","B","A","B"))
# get ids of edges linking v1 and v2
> get.edgeIDs(multi,v=1,alter=2)
[1] 2 1
Notice that to be able to distinguish which edges belonged to each tie type I added a vertex attribute, but this is not a requirement for multiplex edges.
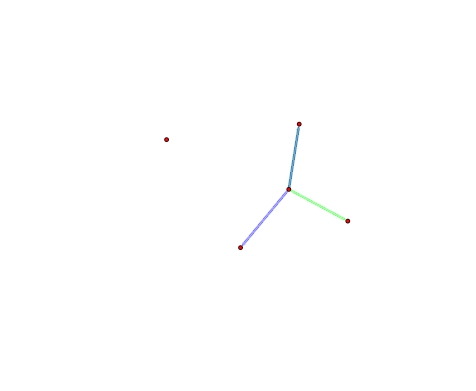
We can plot the multiplex networks, but the plot code basically ignores the multiplex aspect and just treats all the edges the same and draws them on top of eachother. So we have to roll our own coloring function.
# plot the types in different colors with some transparency to show overlap
> plot(multi,edge.col=ifelse(multi%e%'type'=='A',
'#00FF0055','#0000FF55'),edge.lwd=10)
Jacques Philip, a user on the help list, submitted a nice trick to curve the edges so they don’t overlap in the plot (at least when there are just two edge types).
> plot(multi,edge.col=ifelse(multi%e%'type'=='A',
'#00FF0055','#0000FF55'),edge.lwd=10, usecurve=T,
edge.curve=ifelse(multi%e%'type'=='A', 0.3, -0.1))
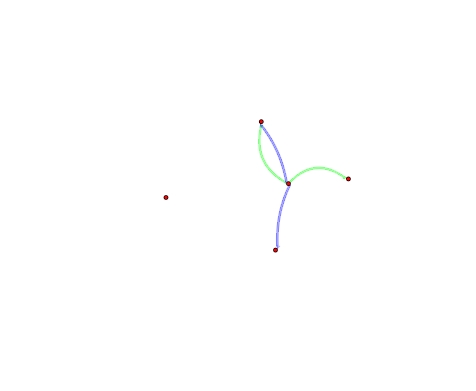
We can use the edge attribute to figure out which edge is each type, and do some crude extraction operations
# get the ids of all edges with type B
> which(get.edge.attribute(multi,'type')=='B')
> [1] 2 4
# create a new network with only type B edges
> multiB< -multi
> multiB< -delete.edges(multiB,
eid=which(get.edge.attribute(multi,'type')!='B'))
> multiB%e%'type'
[1] "B" "B"
> is.multiplex(multiB)
[1] TRUE
Notice that it still says it is a multiplex network, even though we deleted the parallel edges. Its important to remember that is.multiplex() just checks the flag, it doesn’t actually search through the network to see if there are multiplex ties.
Unfortunately, most of the built in functions don’t know what to do with a multiplex network. For example, we can’t print it to a matrix because we don’t have a way of knowing which edge values should be used.
> as.matrix(multi)
Error in as.matrix.network.adjacency(x = x, attrname = attrname, ...) :
Multigraphs not currently supported in as.matrix.network.adjacency. Exiting.
In short, the basic data structures are there, but the multiplex functionality isn’t built out much. Feel free to submit use-cases and feature requests.
P.S. For most situations you probably don’t want to use multiplex tie types to represent multiple waves of networks. The networkDynamic package provides better structures for that.
P.P.S. Now if I could just get WordPress not to mangle R’s "< -" assignment operator so that it turns into "lessthan minus"

Thanks for this. The documentation of statnet sometimes drives me crazy, but posts like this are useful.
Hi This is useful. I wonder if the ergm can be made to deal with this? I want to use an edge covariate for multiple transactions in a trader network but I get the Error in as.matrix.network.adjacency(x = x, attrname = attrname, …) :
Multigraphs not currently supported in as.matrix.network.adjacency. Exiting.
>
I imagine that a response a year late is not very helpful, but have to posted to the statnet help mailing list? https://statnet.csde.washington.edu/statnet_users_group.shtml
Hi, I am considering simulating a network with nodes having two different types of edge (for example direct and indirect contact). I was wondering if there is a way to specify the formation and target stats the same way as one would do for a regular network with single edge type?